Discover SAMSON, your integrative
platform for molecular design
Turn on the sound of the video
for the best experience
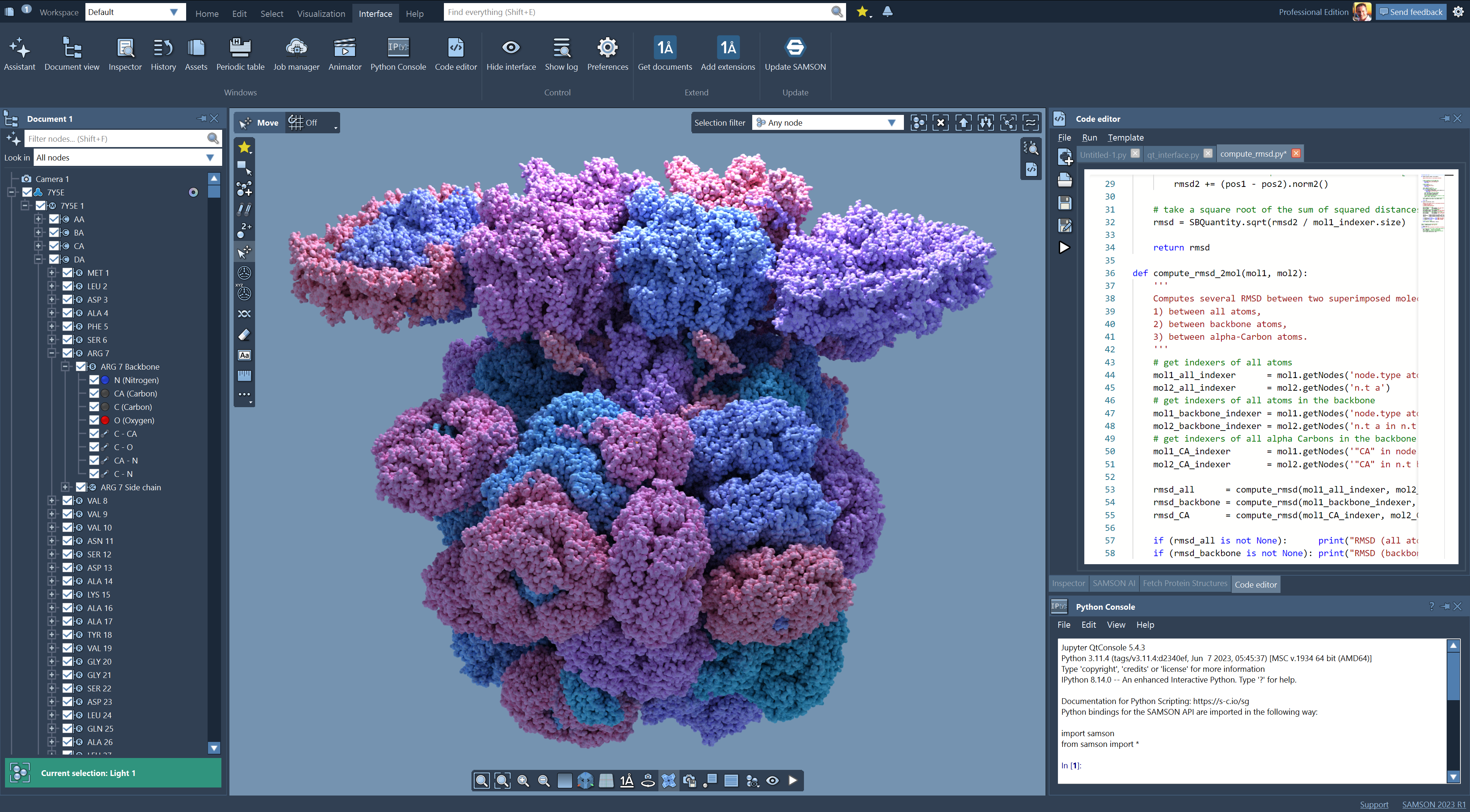
Meet SAMSON
SAMSON is the quickly growing platform for integrated molecular
design.
SAMSON's goal is to make it faster for everyone to design drugs, materials and nanosystems.
For fun, or to build a better tomorrow through science and innovation in medicine, energy, materials, electronics, education, and more.
SAMSON is available on all major operating systems.
Learn more.
SAMSON's goal is to make it faster for everyone to design drugs, materials and nanosystems.
For fun, or to build a better tomorrow through science and innovation in medicine, energy, materials, electronics, education, and more.
SAMSON is available on all major operating systems.
Learn more.
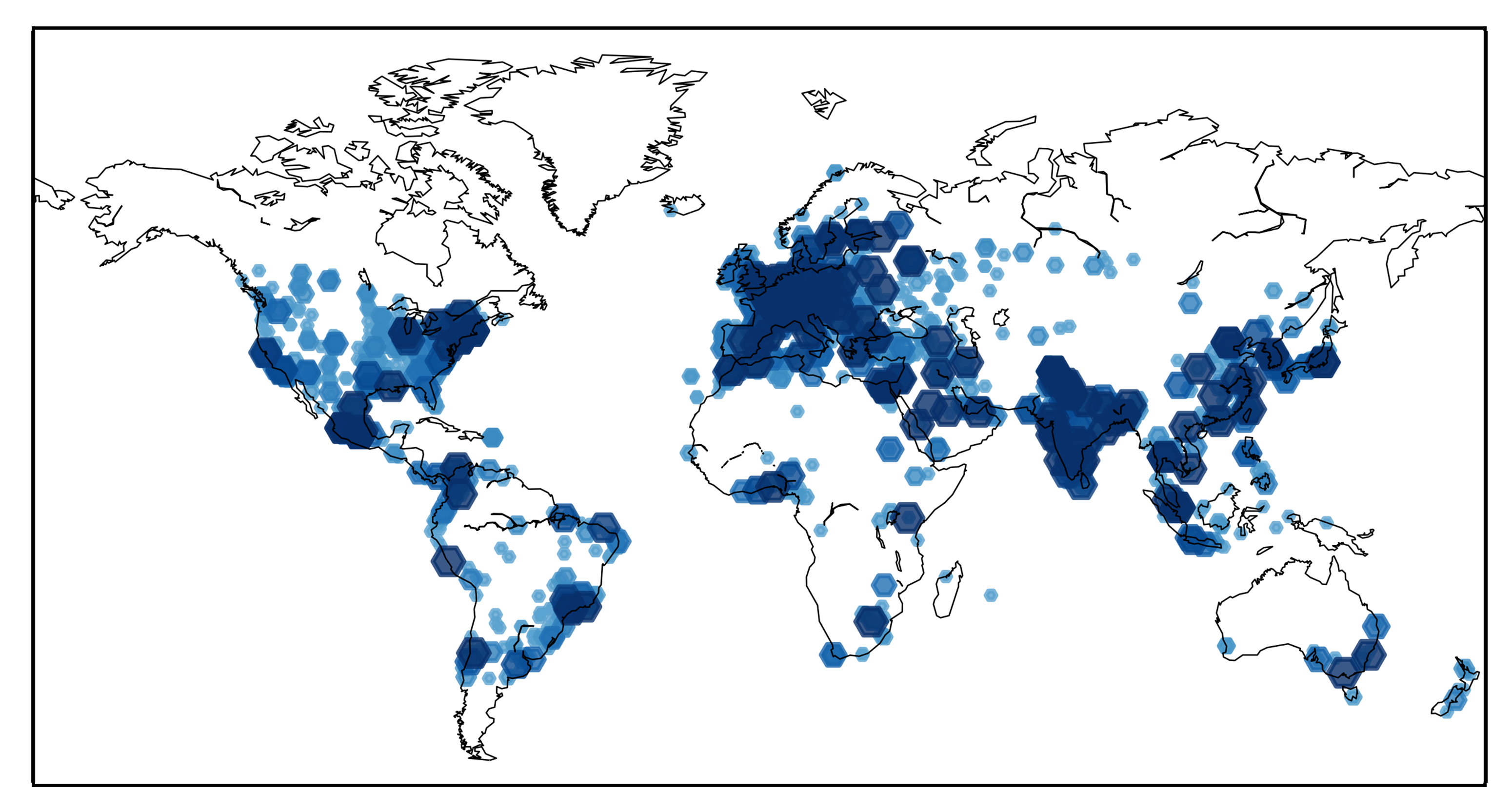
Meet the community
SAMSON has thousands of registered users and developers,
both in academia and industry.
Network and find collaborators for your projects.
Share tips and tricks.
Exchange SAMSON documents, models, calculations, scripts, references, papers, and more.
Learn more.
both in academia and industry.
Network and find collaborators for your projects.
Share tips and tricks.
Exchange SAMSON documents, models, calculations, scripts, references, papers, and more.
Learn more.
Platform
+
Countries
+
Labs and Companies
+
Extensions Installed
A modern, feature-packed
platform for molecular design
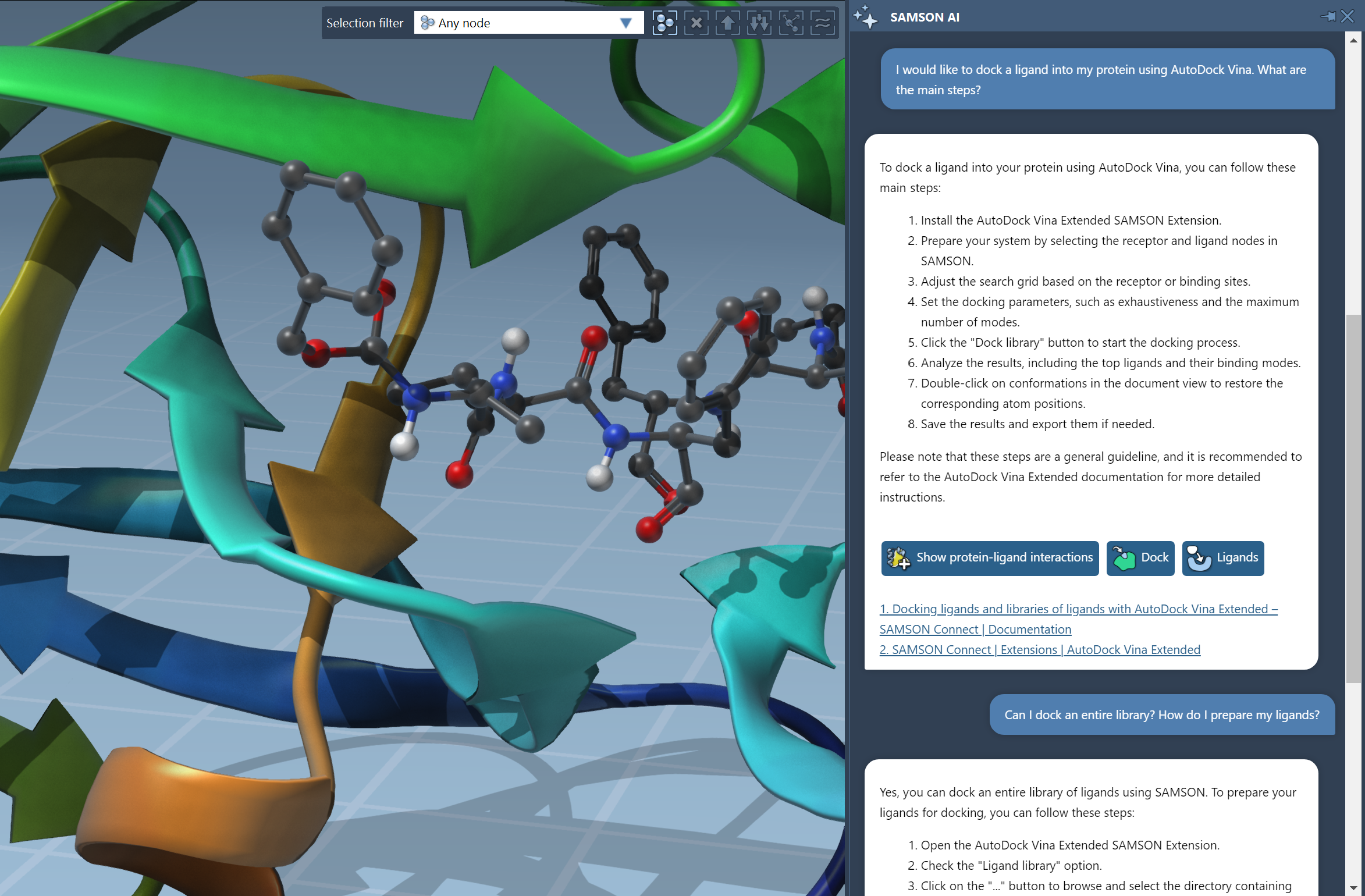
Your AI Assistant
Ask SAMSON AI for help with molecular design and get personalized
answers.
Get direct access to the exact commands and documentation pages you need, streamlining your workflow like never before.
Make SAMSON AI read web pages and online PDFs, and ask it questions about them.
Ask SAMSON AI for help when writing Python scripts and creating Apps.
Use AI commands to make SAMSON work for you.
Control SAMSON AI with your voice instead of typing.
Learn more.
Get direct access to the exact commands and documentation pages you need, streamlining your workflow like never before.
Make SAMSON AI read web pages and online PDFs, and ask it questions about them.
Ask SAMSON AI for help when writing Python scripts and creating Apps.
Use AI commands to make SAMSON work for you.
Control SAMSON AI with your voice instead of typing.
Learn more.
Build and simulate.
At the same time.
At the same time.
Quickly build complex models by assembling atoms, fragments and
assets.
Use physics-based construction to create molecules and assemblies.
Model nanosystems through their structures, dynamics, interactions, visuals, and properties.
Enable expert-in-the-loop design with interactive simulations.
Export to large-scale simulators, and import trajectories to compute properties.
Learn more.
Use physics-based construction to create molecules and assemblies.
Model nanosystems through their structures, dynamics, interactions, visuals, and properties.
Enable expert-in-the-loop design with interactive simulations.
Export to large-scale simulators, and import trajectories to compute properties.
Learn more.
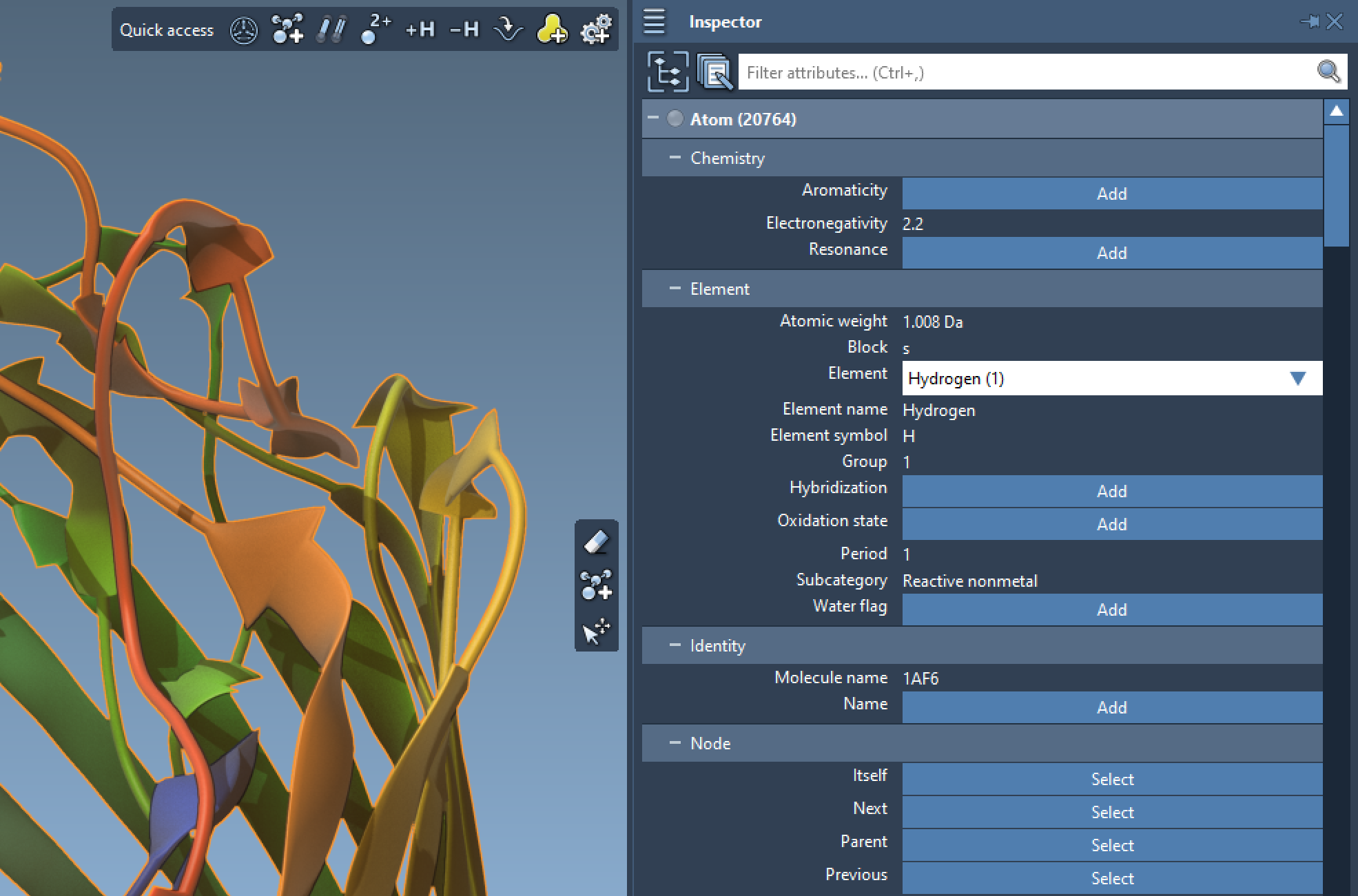
Control
View and edit all attributes with the inspector.
Control what you edit: the last selected node, or all selected nodes simultaneously.
Monitor interactive simulations.
Add missing properties with complete control.
Learn more.
Control what you edit: the last selected node, or all selected nodes simultaneously.
Monitor interactive simulations.
Add missing properties with complete control.
Learn more.
Stay in the zone
Save time with a modern viewport which lets you do more.
Optimize your workflow with contextual Quick Access commands.
Control your selections thanks to the advanced Selection Filter.
Activate your favorite apps and editors in one click.
Orient the view with the Compass.
Learn more.
Optimize your workflow with contextual Quick Access commands.
Control your selections thanks to the advanced Selection Filter.
Activate your favorite apps and editors in one click.
Orient the view with the Compass.
Learn more.
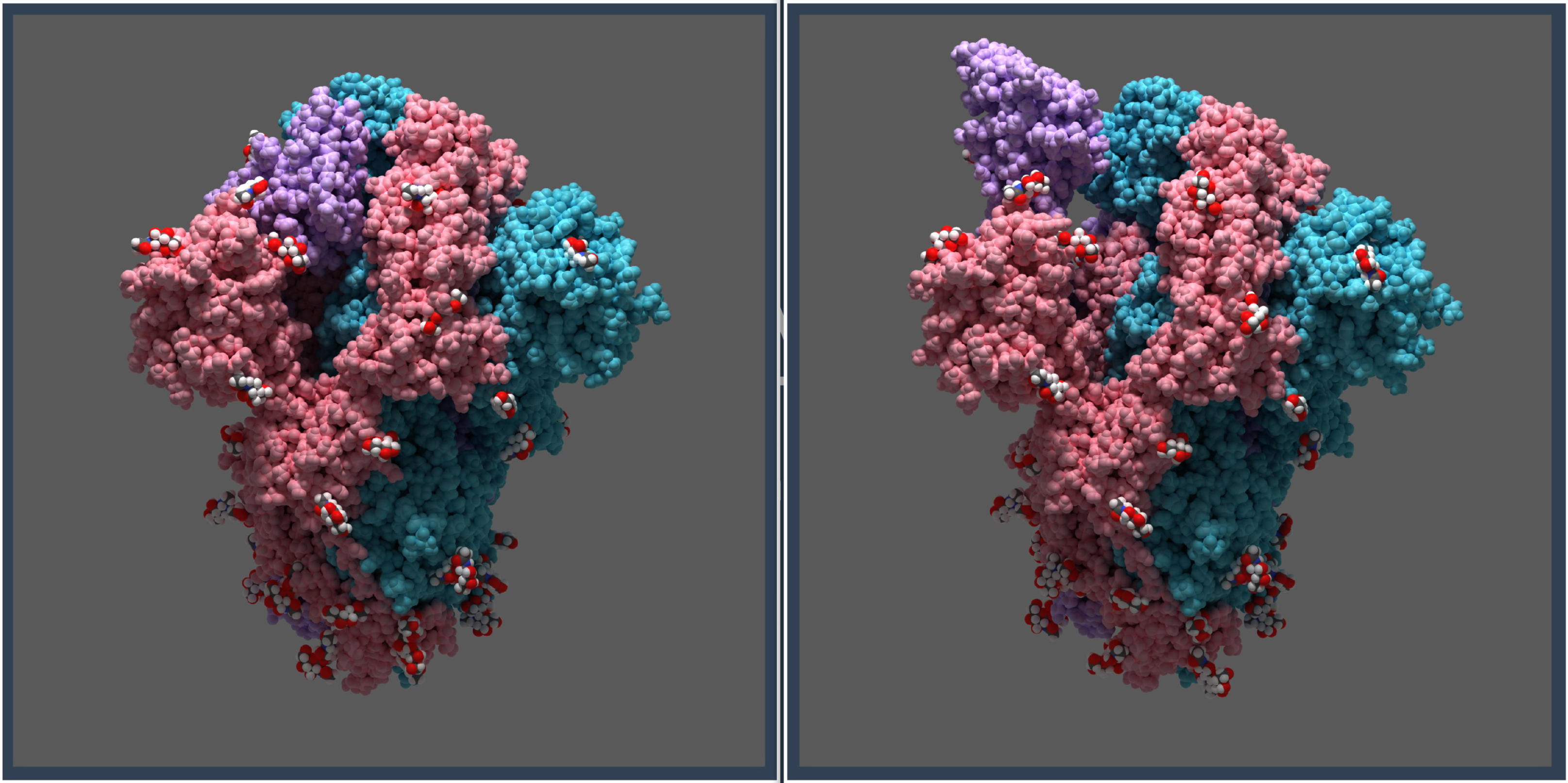
Biology ready
Fetch structures from databases or use protein structure
prediction to build new models.
Quickly select receptors, ligands, binding sites, waters, ions, sequences, etc., and perform advanced selections.
Control alternate positions, non-standard residues and clashes with the structure validation tool.
Apply custom visual representations and color schemes.
Mutate residues and edit rotamers.
Learn more.
Quickly select receptors, ligands, binding sites, waters, ions, sequences, etc., and perform advanced selections.
Control alternate positions, non-standard residues and clashes with the structure validation tool.
Apply custom visual representations and color schemes.
Mutate residues and edit rotamers.
Learn more.
A unique platform for
science communication
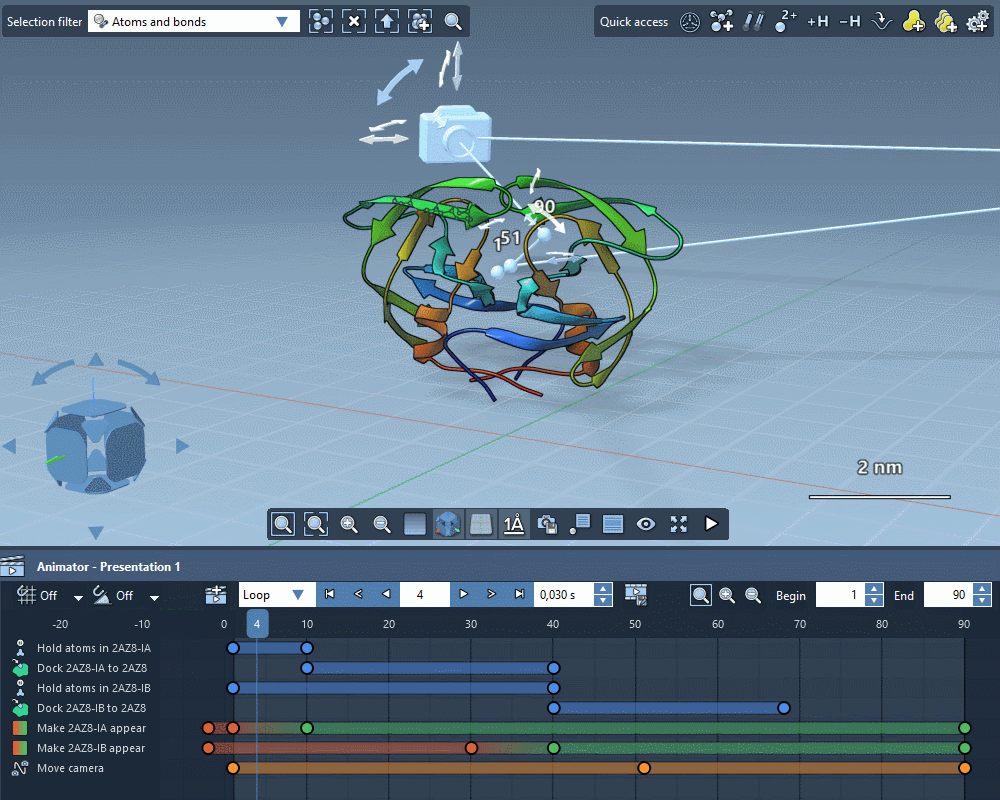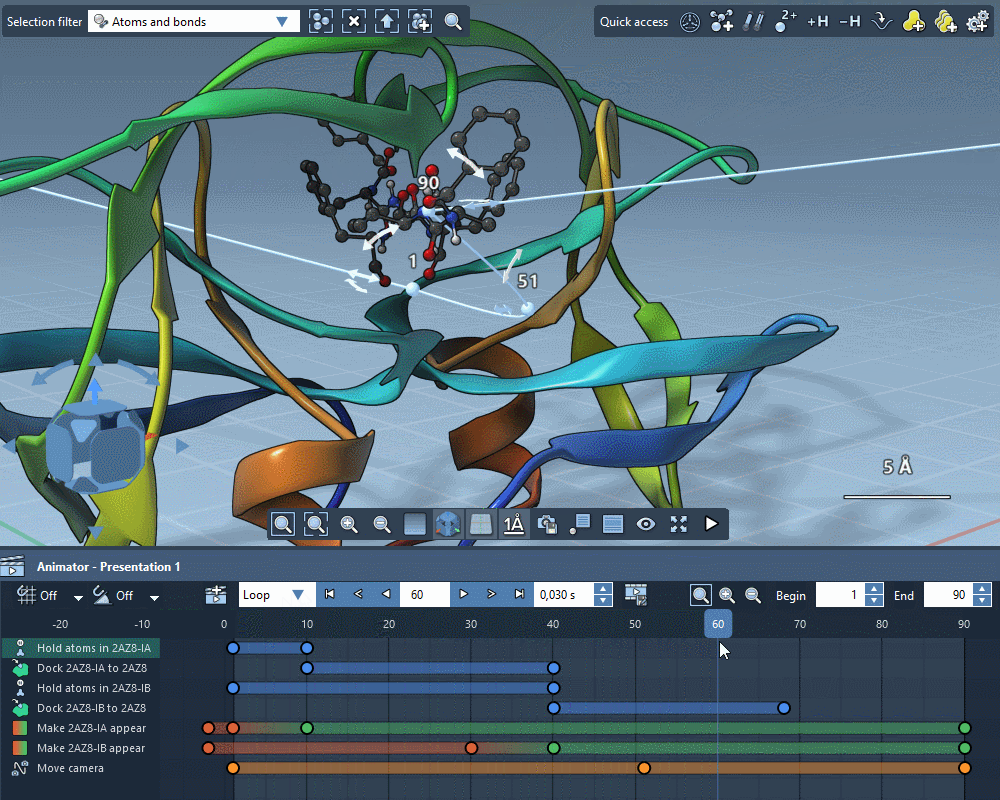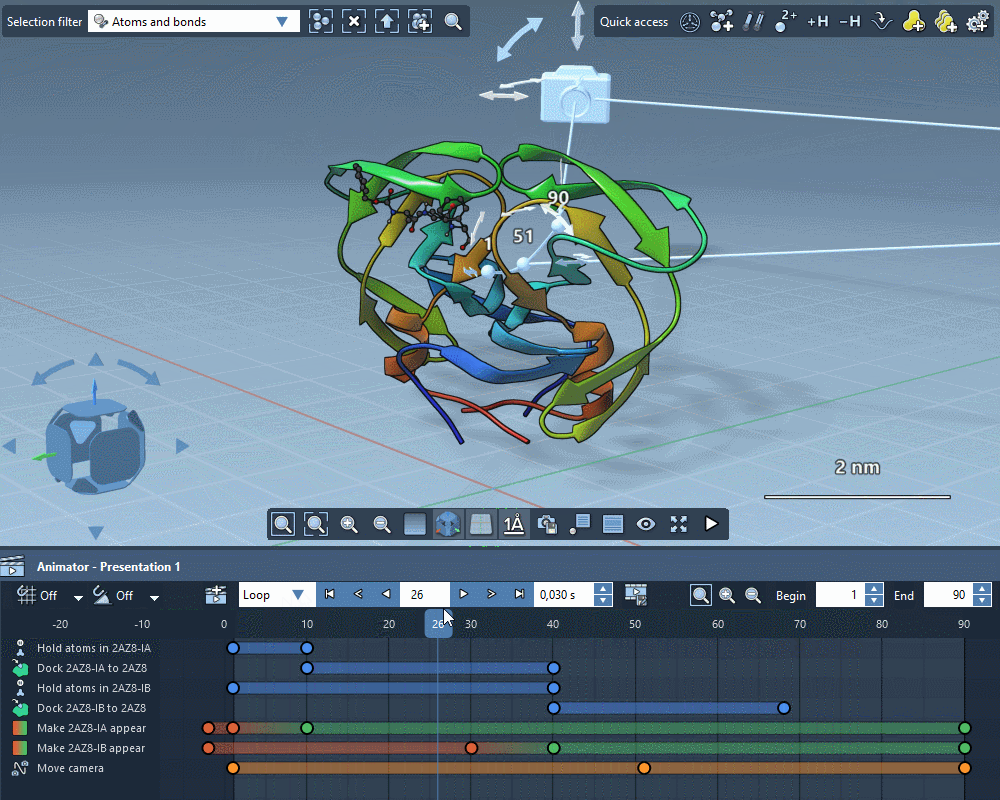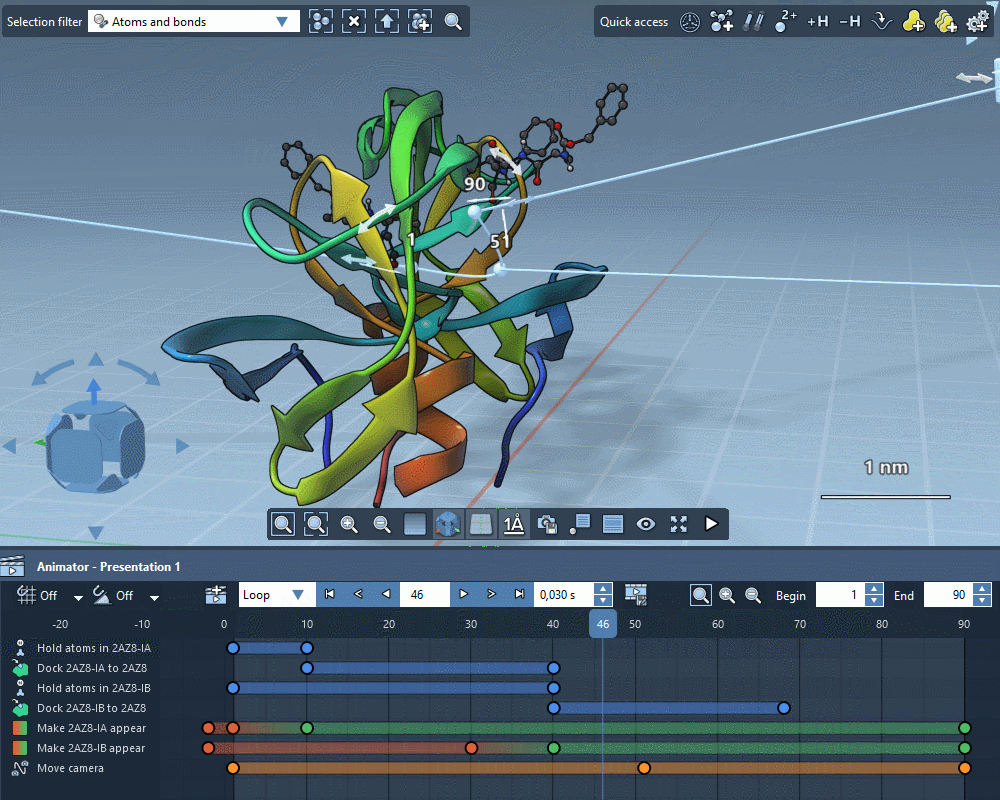
Animate everything
Create gorgeous animations with the SAMSON Animator.
Use templates to quickly animate structures and cameras.
Explain molecular mechanisms thanks to structure-aware animations.
Add animation effects in a few clicks.
Learn more.
Use templates to quickly animate structures and cameras.
Explain molecular mechanisms thanks to structure-aware animations.
Add animation effects in a few clicks.
Learn more.
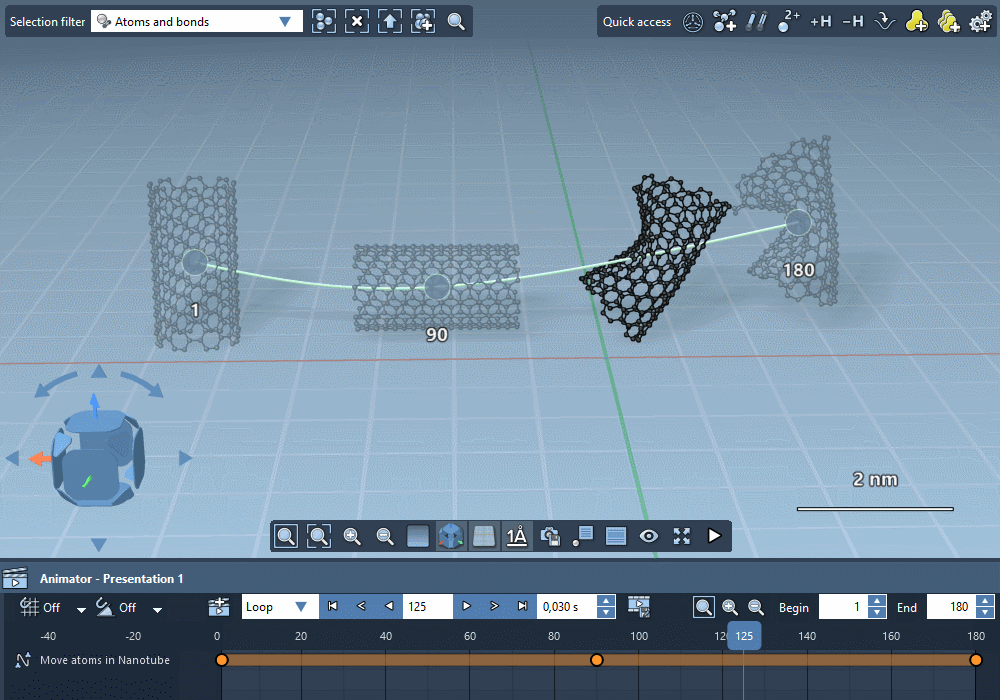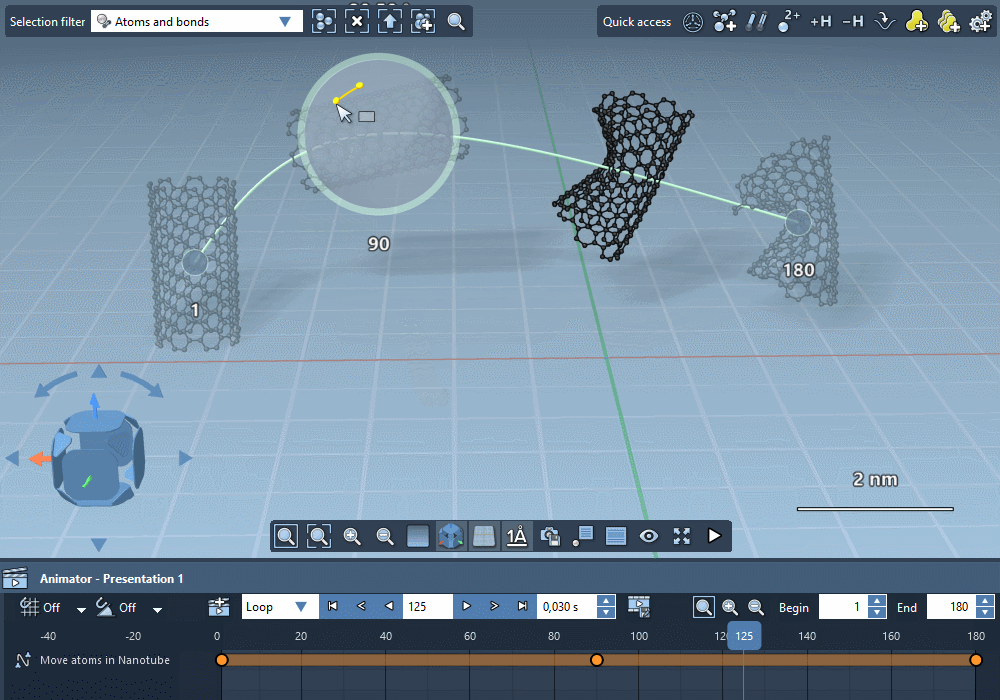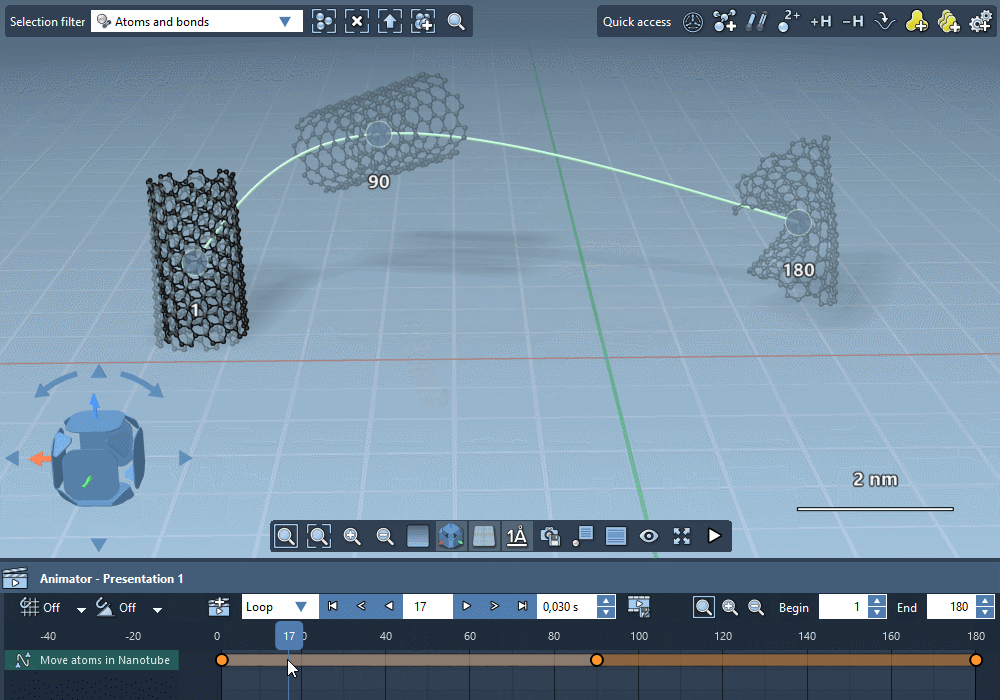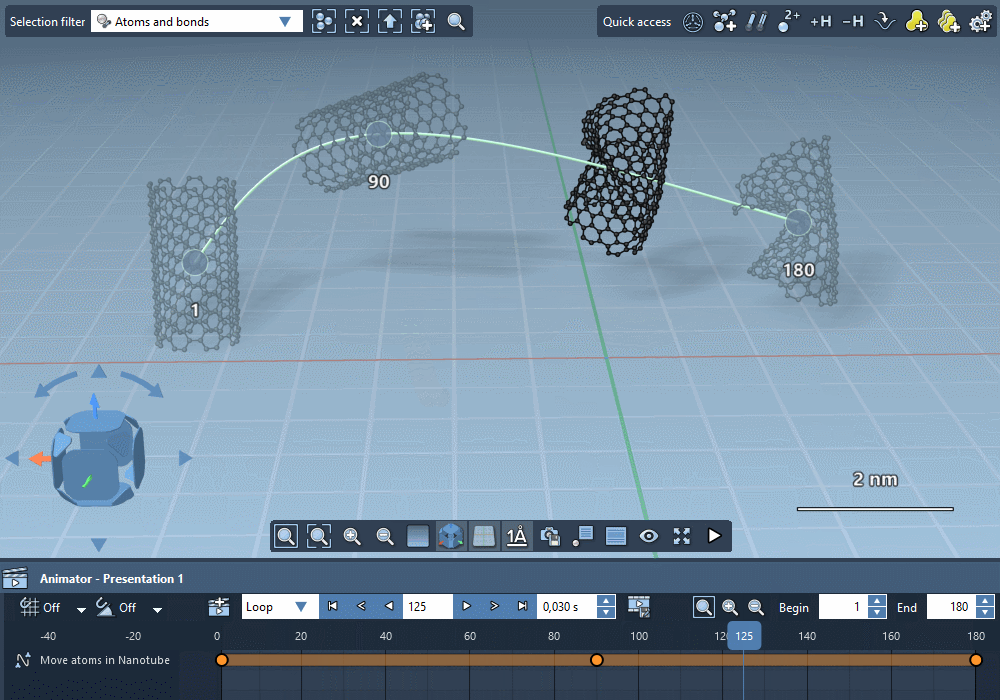
Integrate trajectories.
Or create them.
Or create them.
Import simulation trajectories to integrate them in your
animations.
Integrate paths computed by SAMSON extensions.
Create custom trajectories using controllable keyframes.
Learn more.
Integrate paths computed by SAMSON extensions.
Create custom trajectories using controllable keyframes.
Learn more.
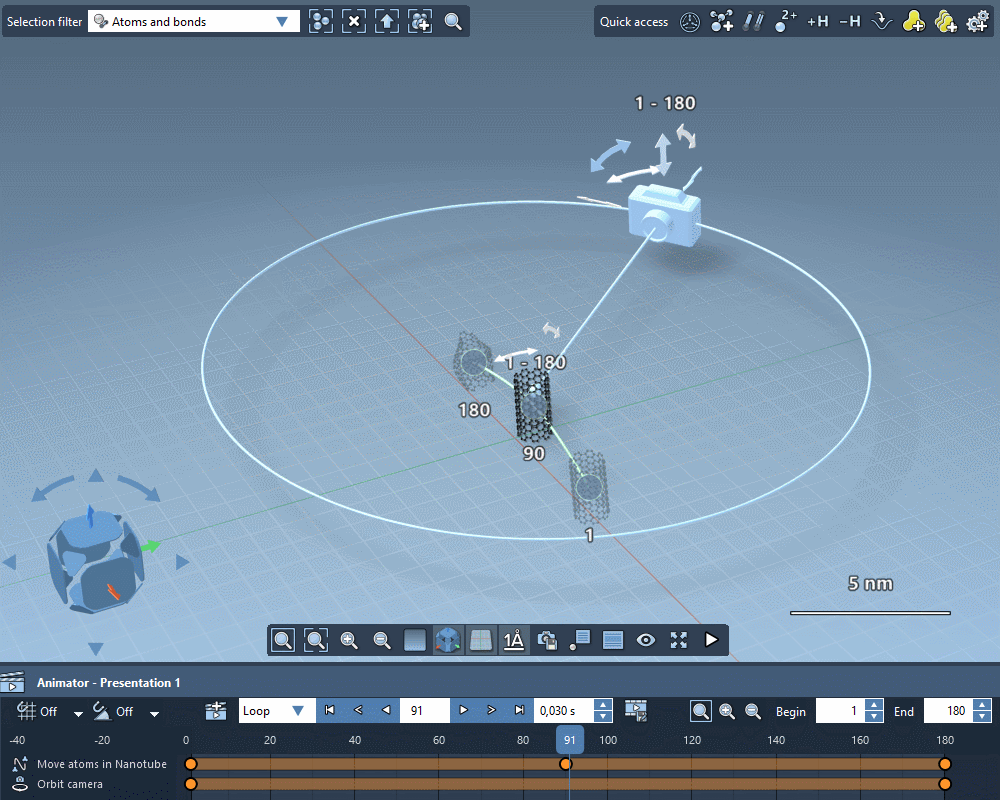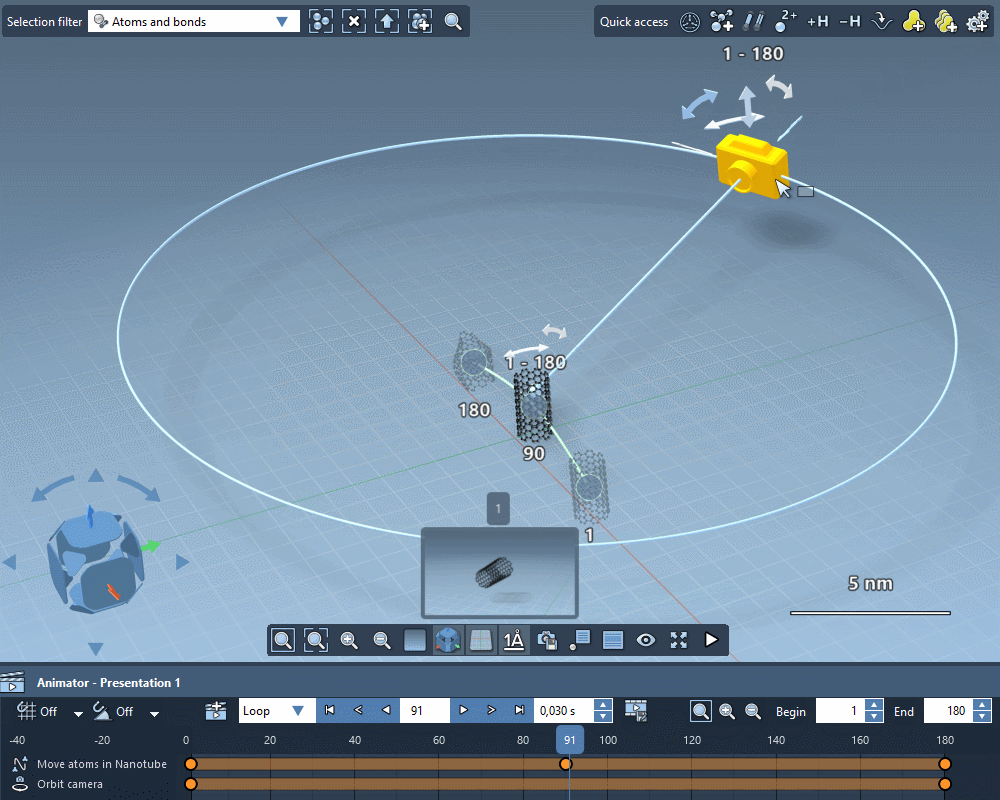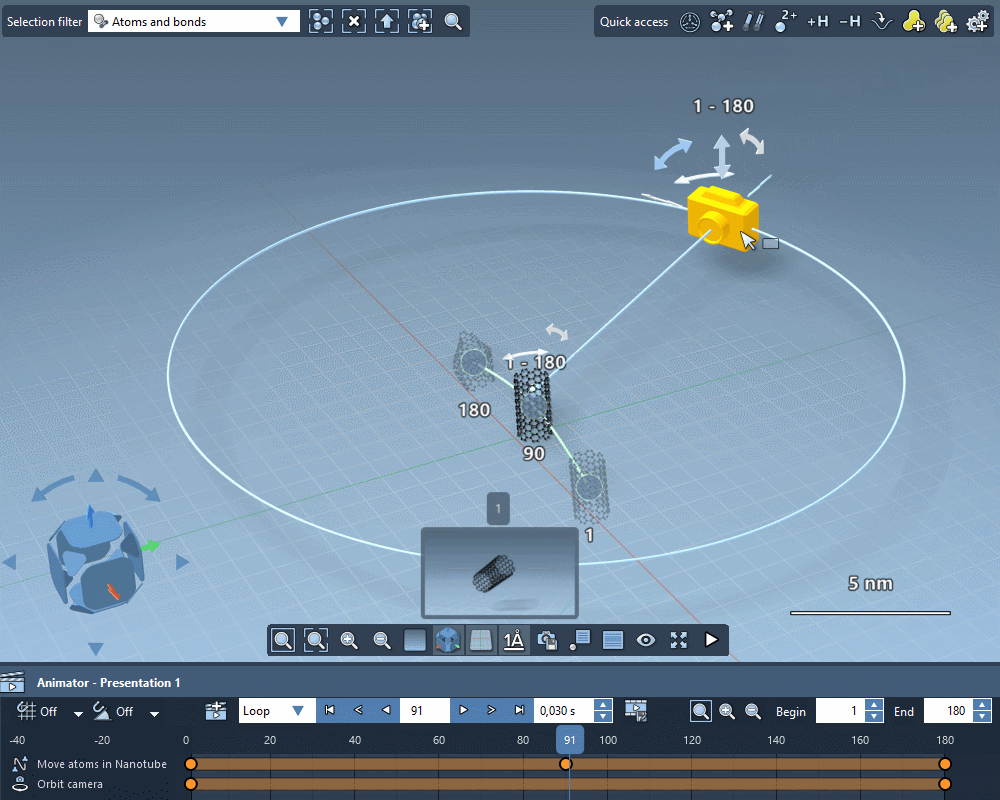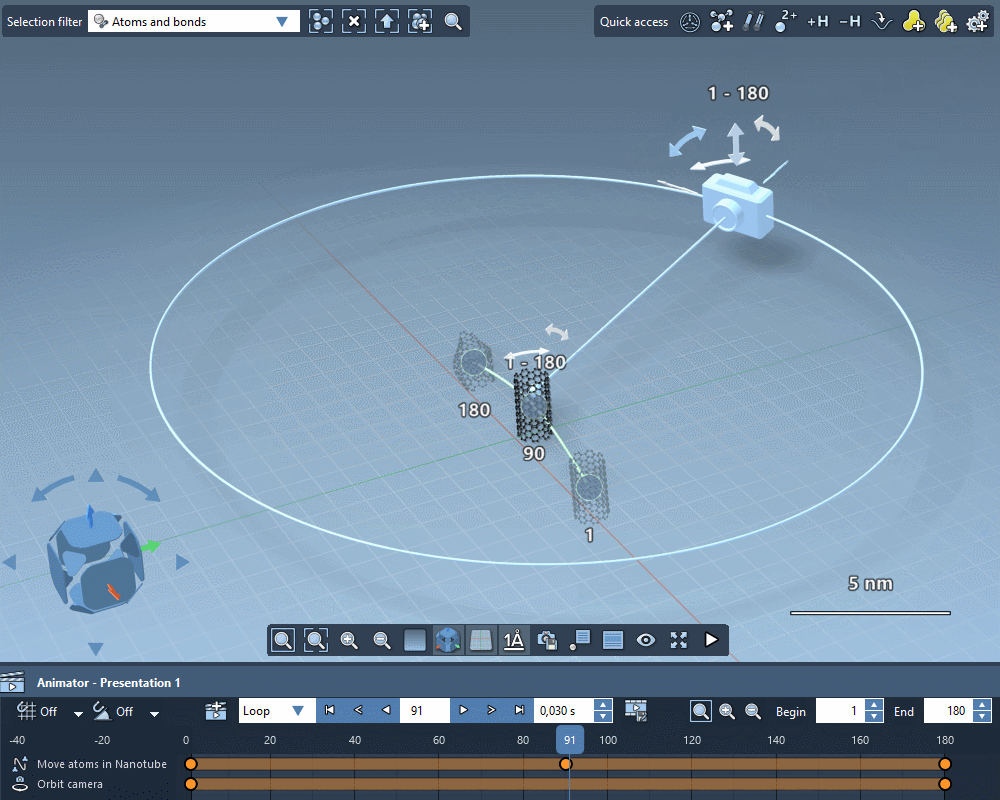
Take the perfect shot
Quickly apply standard camera motions to your animations.
Create custom camera motions using keyframes.
Add intelligent cameras that automatically follow atoms selections.
Learn more.
Create custom camera motions using keyframes.
Add intelligent cameras that automatically follow atoms selections.
Learn more.
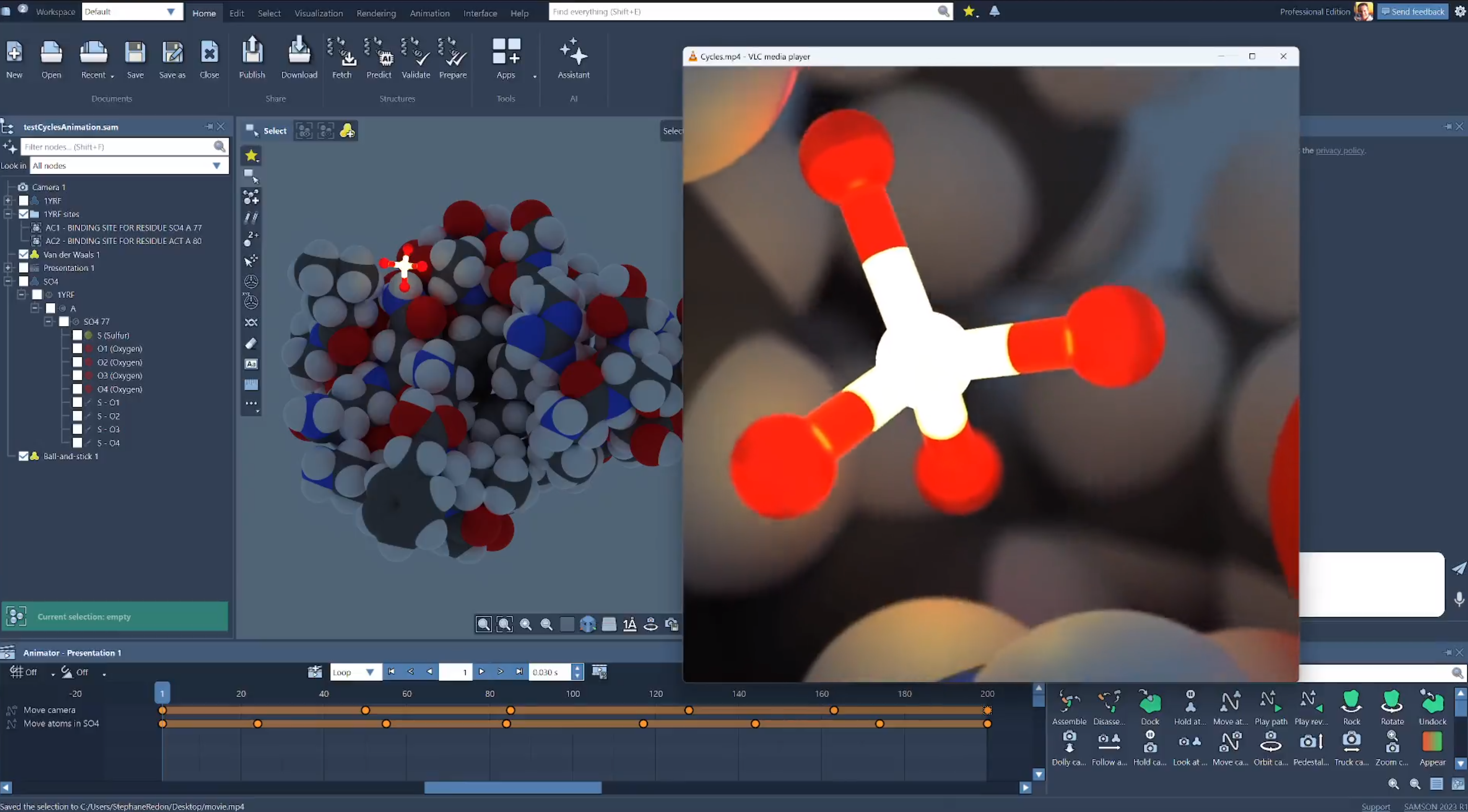
Render
Use custom visual representations to highlight features and
communicate your results.
Apply visual presets and render presets to create advanced visualizations in a few clicks.
Produce publication-quality images with advanced rendering options, including ambient occlusion, bloom, depth of field, silhouettes, etc.
Unleash your creativity in one click with Cycles, the famed path tracer used in animation studios, and create photorealistic renderings.
Share your creations on SAMSON Connect.
Learn more.
Apply visual presets and render presets to create advanced visualizations in a few clicks.
Produce publication-quality images with advanced rendering options, including ambient occlusion, bloom, depth of field, silhouettes, etc.
Unleash your creativity in one click with Cycles, the famed path tracer used in animation studios, and create photorealistic renderings.
Share your creations on SAMSON Connect.
Learn more.
A platform that helps you
achieve more
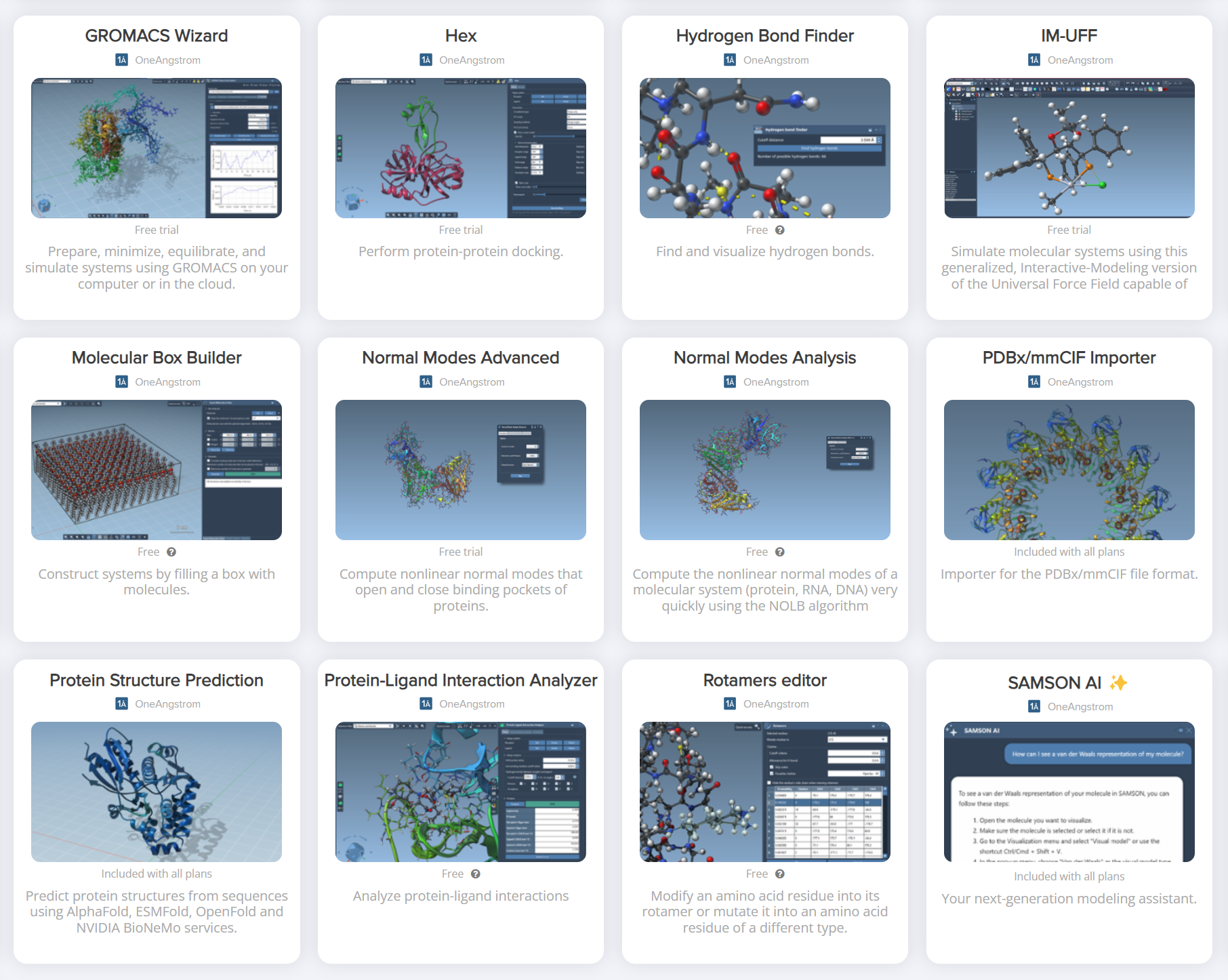
Extend SAMSON
SAMSON is an open platform, and that's what makes it so different
and powerful.
Head to the marketplace for SAMSON Extensions to dock ligands into proteins, analyze experimental images of graphene, perform molecular dynamics, immerse yourself in virtual reality, etc.
Combine SAMSON Extensions together to build your own workflow.
Access state-of-the-art algorithms from academia and industry.
Add free SAMSON Extensions in one click or subscribe to paid SAMSON Extensions.
All added extensions are automatically installed when you restart your SAMSON.
Learn more.
Head to the marketplace for SAMSON Extensions to dock ligands into proteins, analyze experimental images of graphene, perform molecular dynamics, immerse yourself in virtual reality, etc.
Combine SAMSON Extensions together to build your own workflow.
Access state-of-the-art algorithms from academia and industry.
Add free SAMSON Extensions in one click or subscribe to paid SAMSON Extensions.
All added extensions are automatically installed when you restart your SAMSON.
Learn more.

Embed files
Embed any number of files - whatever their types - and folders in
your SAMSON documents.
Include PDFs, research notes, molecular libraries - even other SAMSON documents - and all files that are relevant to your work to make it easy to share your research with the world.
Learn more.
Include PDFs, research notes, molecular libraries - even other SAMSON documents - and all files that are relevant to your work to make it easy to share your research with the world.
Learn more.
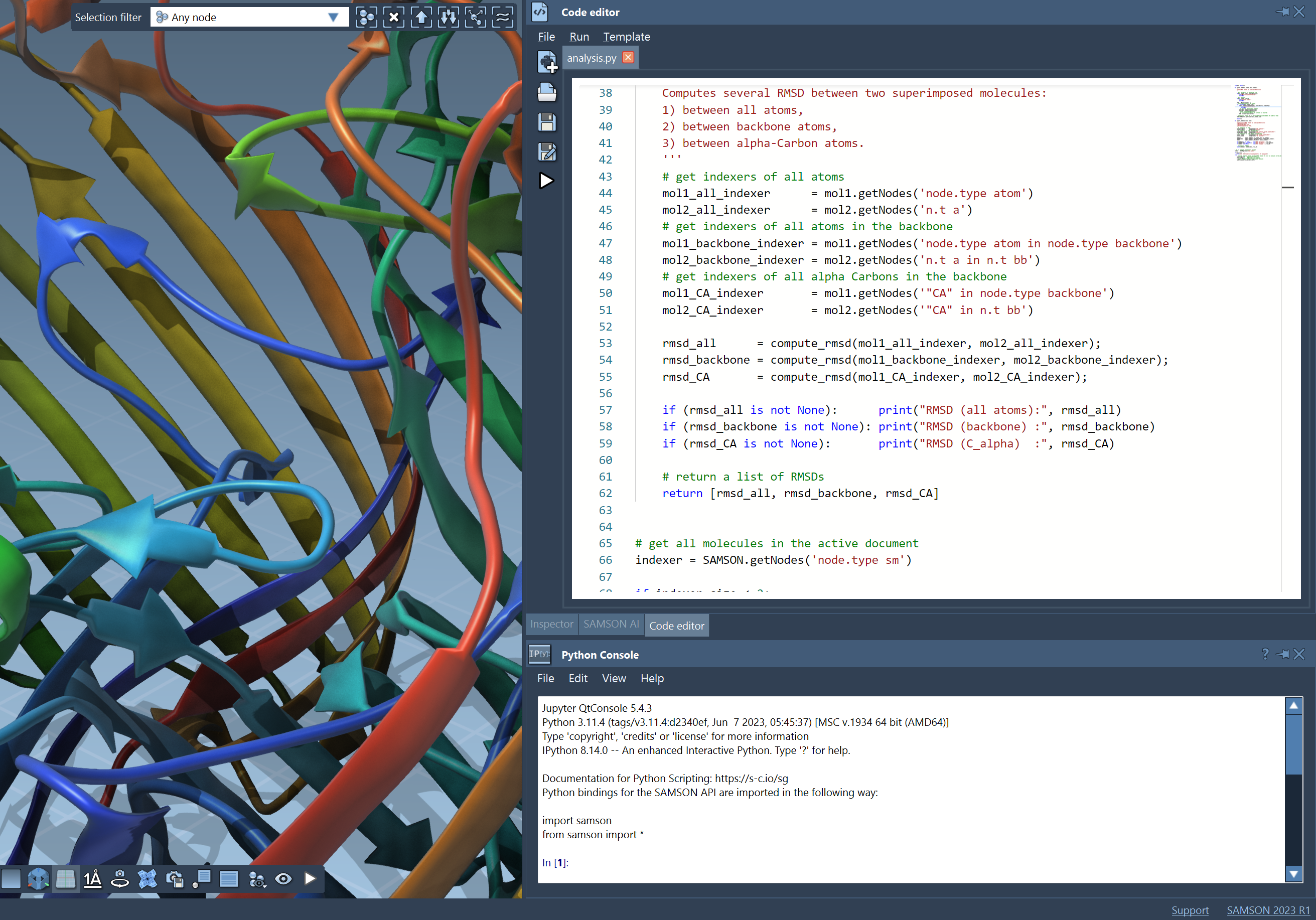
Script
Use the integrated Python interpreter, editor and console to
rapidly develop scripts and apps for molecular design.
Pipeline calculations.
Integrate packages for deep learning, protein design, chemistry, statistics, and more.
Embed your apps in SAMSON documents in a few clicks to create executable documents.
Share your apps with the world on SAMSON Connect.
Learn more.
Pipeline calculations.
Integrate packages for deep learning, protein design, chemistry, statistics, and more.
Embed your apps in SAMSON documents in a few clicks to create executable documents.
Share your apps with the world on SAMSON Connect.
Learn more.

Compute in the cloud
When you need power, use Computing Credits to perform calculations
in the cloud through select extensions.
Create jobs from SAMSON.
Execute them in the cloud.
Get results back into SAMSON.
Refer friends and colleagues to the SAMSON Cloud Services to earn free Computing Credits.
Would you like to learn more?
Schedule a short call.
Create jobs from SAMSON.
Execute them in the cloud.
Get results back into SAMSON.
Refer friends and colleagues to the SAMSON Cloud Services to earn free Computing Credits.
Would you like to learn more?
Schedule a short call.
Friendly with beginners and
experts alike
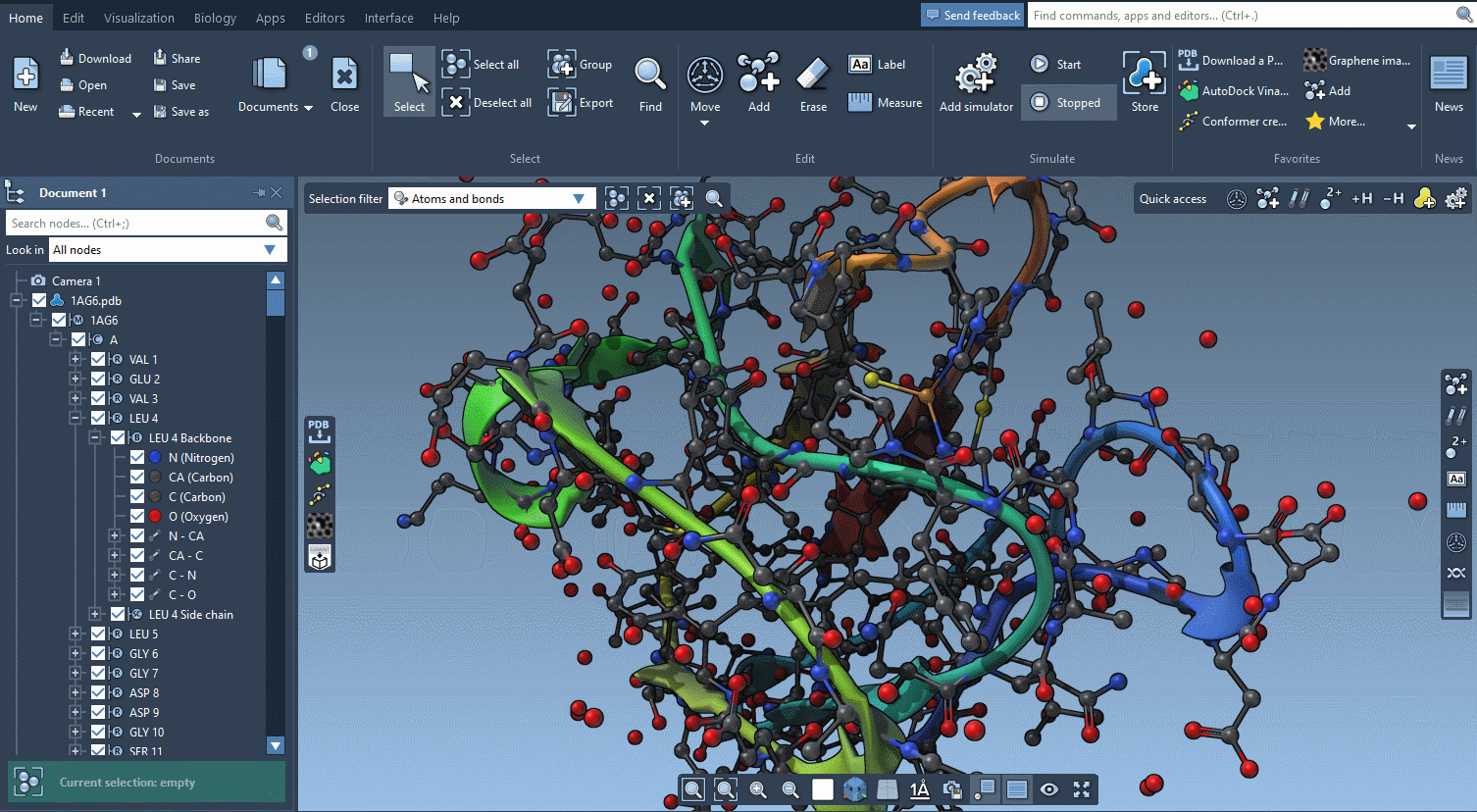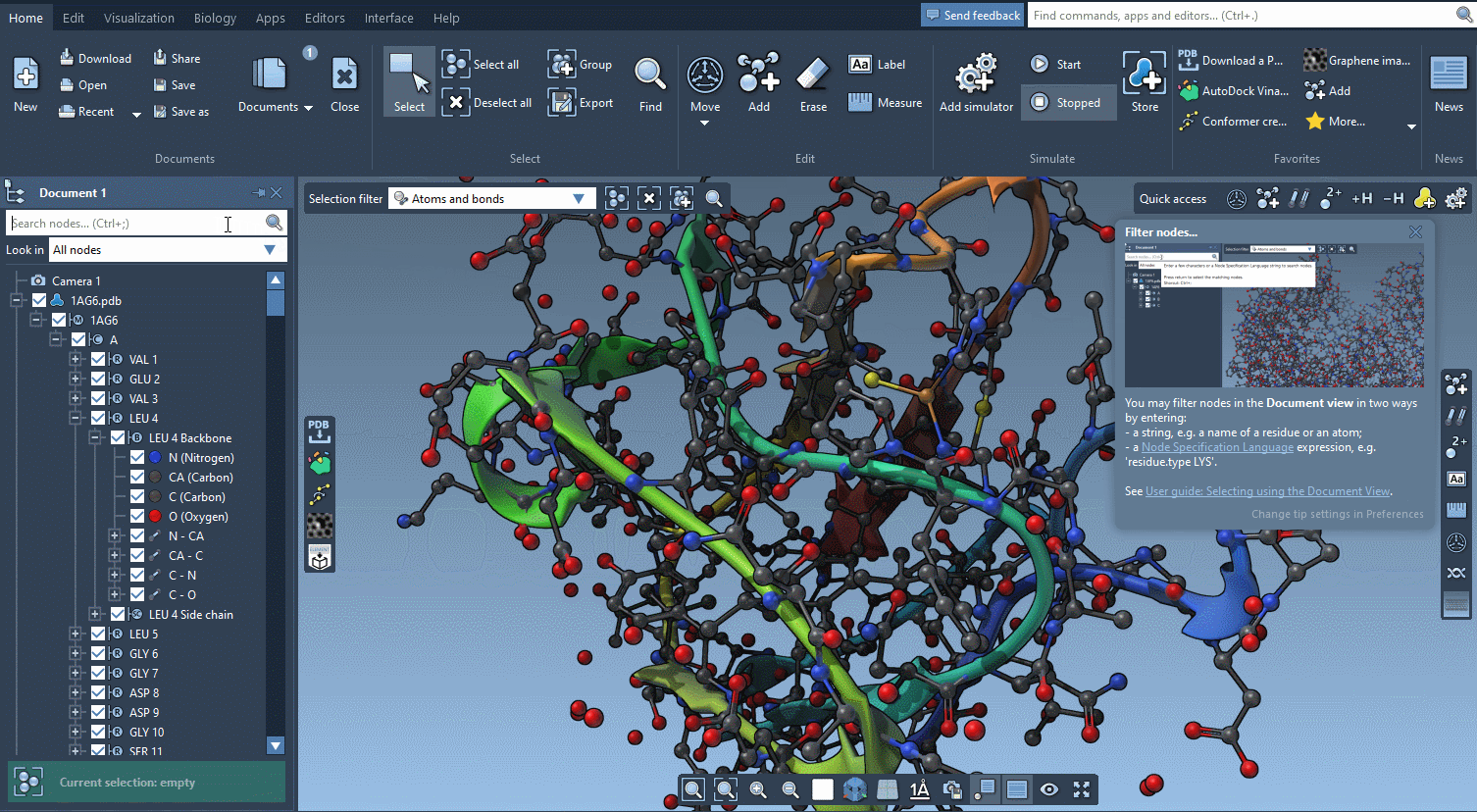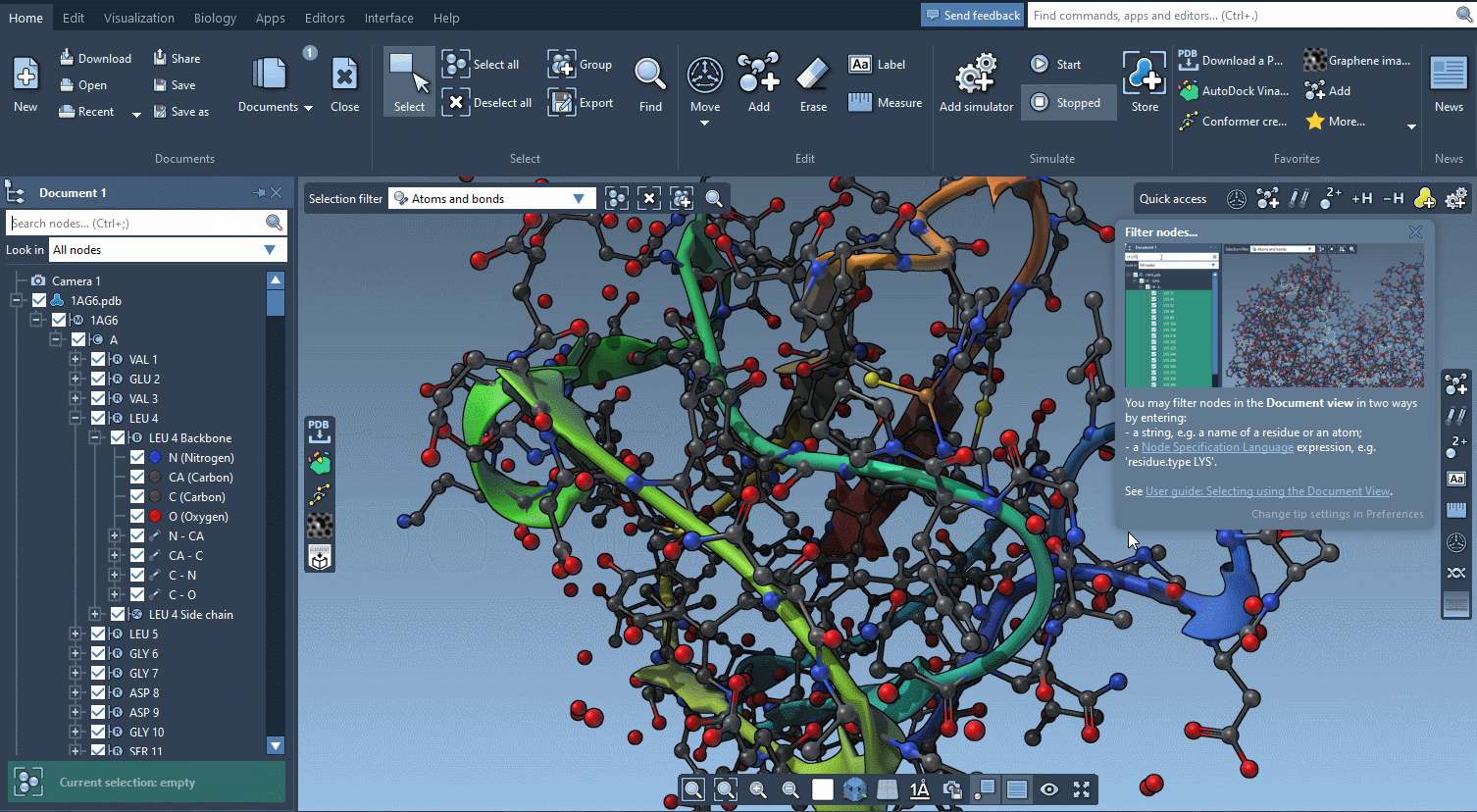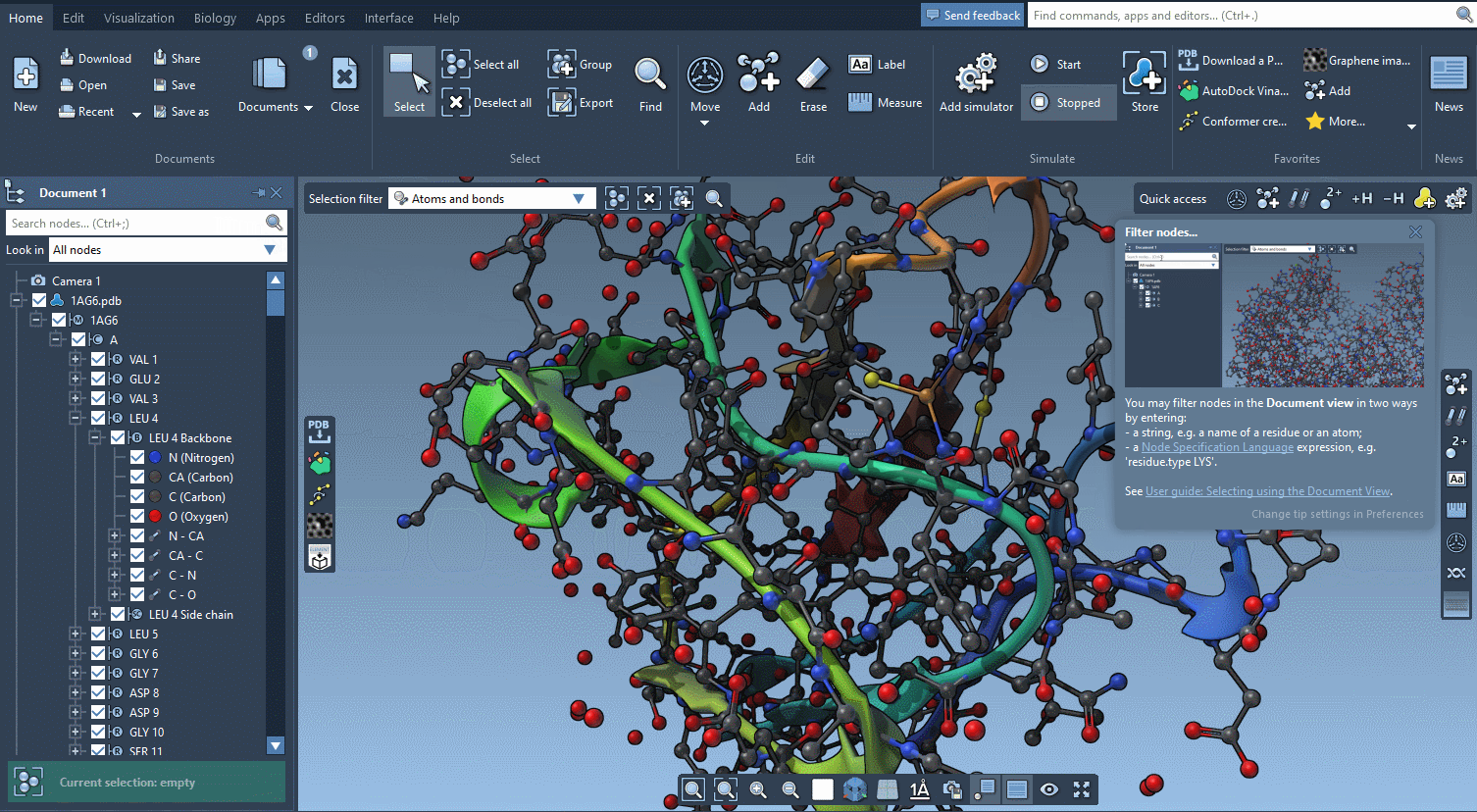
Help included
SAMSON AI has your back, but there is so much more help available.
SAMSON features contextual tips that pop up when you need them.
Start searching for nodes and SAMSON will tell you about the Node Specification Language.
Begin to move atoms and you will learn everything about motion widgets.
SAMSON also comes with guided step-by-step tutorials.
From learning to work with documents and making publication-quality images to performing advanced interactive simulations, quickly get up to speed while learning at your own pace.
Learn more.
SAMSON features contextual tips that pop up when you need them.
Start searching for nodes and SAMSON will tell you about the Node Specification Language.
Begin to move atoms and you will learn everything about motion widgets.
SAMSON also comes with guided step-by-step tutorials.
From learning to work with documents and making publication-quality images to performing advanced interactive simulations, quickly get up to speed while learning at your own pace.
Learn more.
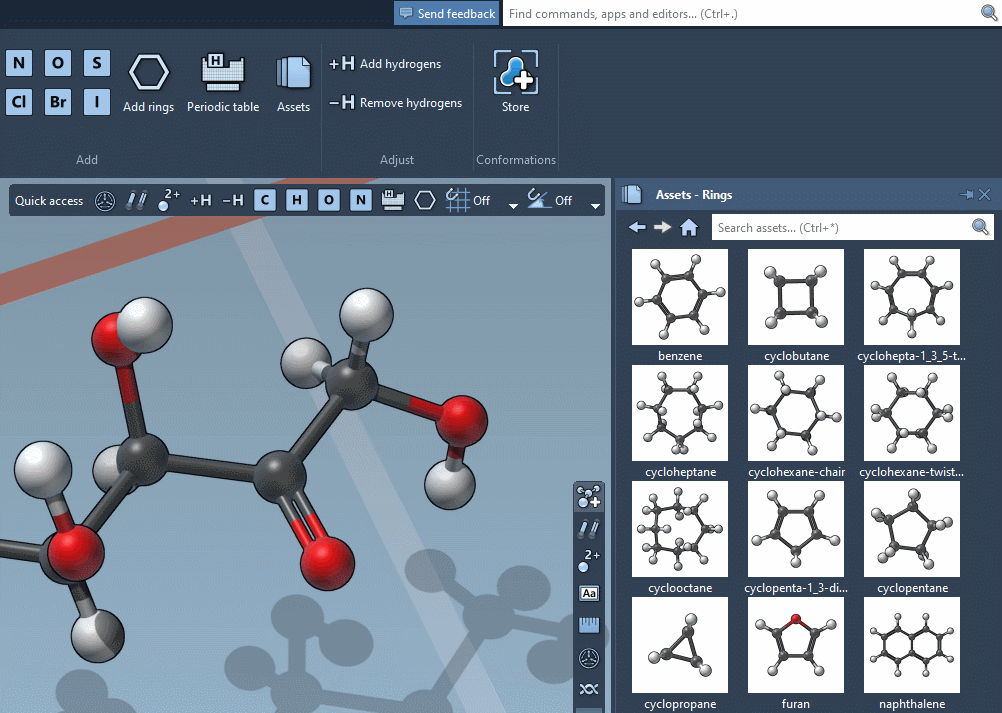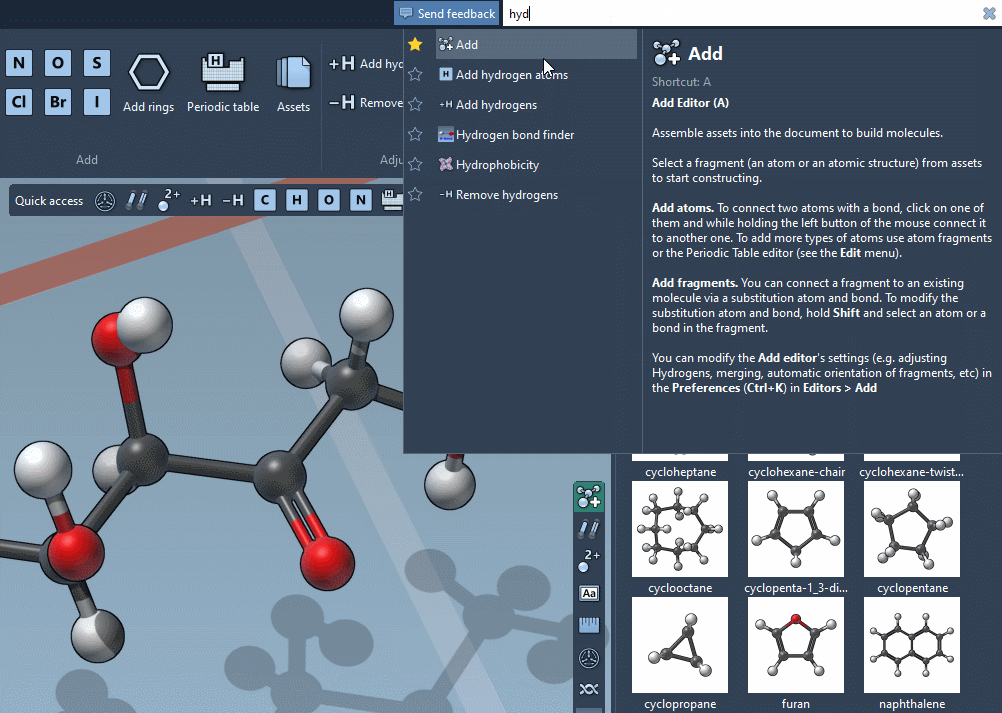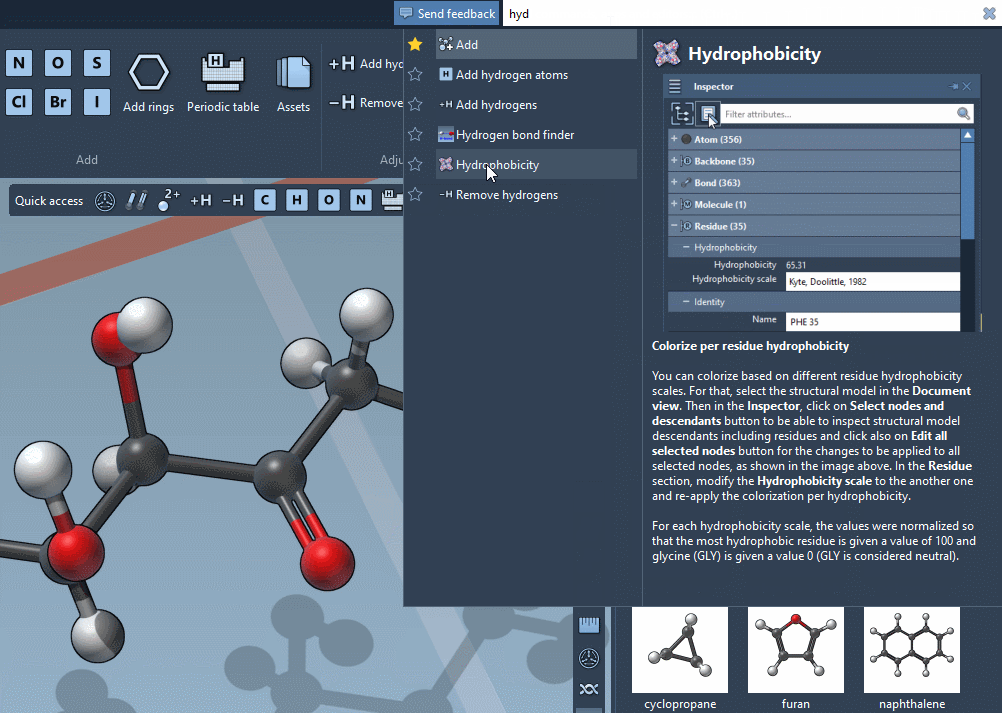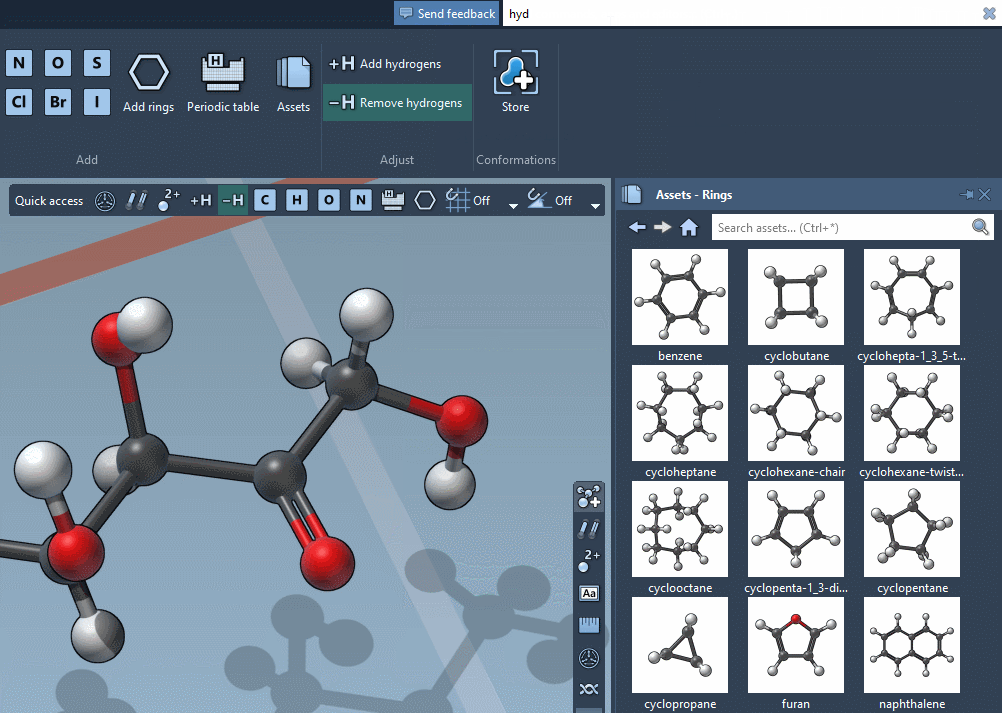
Find everything
Use the Command Finder to instantly find commands, apps, editors,
and more.
Get help snippets and shortcuts reminders.
Favorite your preferred commands for rapid access.
Learn more.
Get help snippets and shortcuts reminders.
Favorite your preferred commands for rapid access.
Learn more.
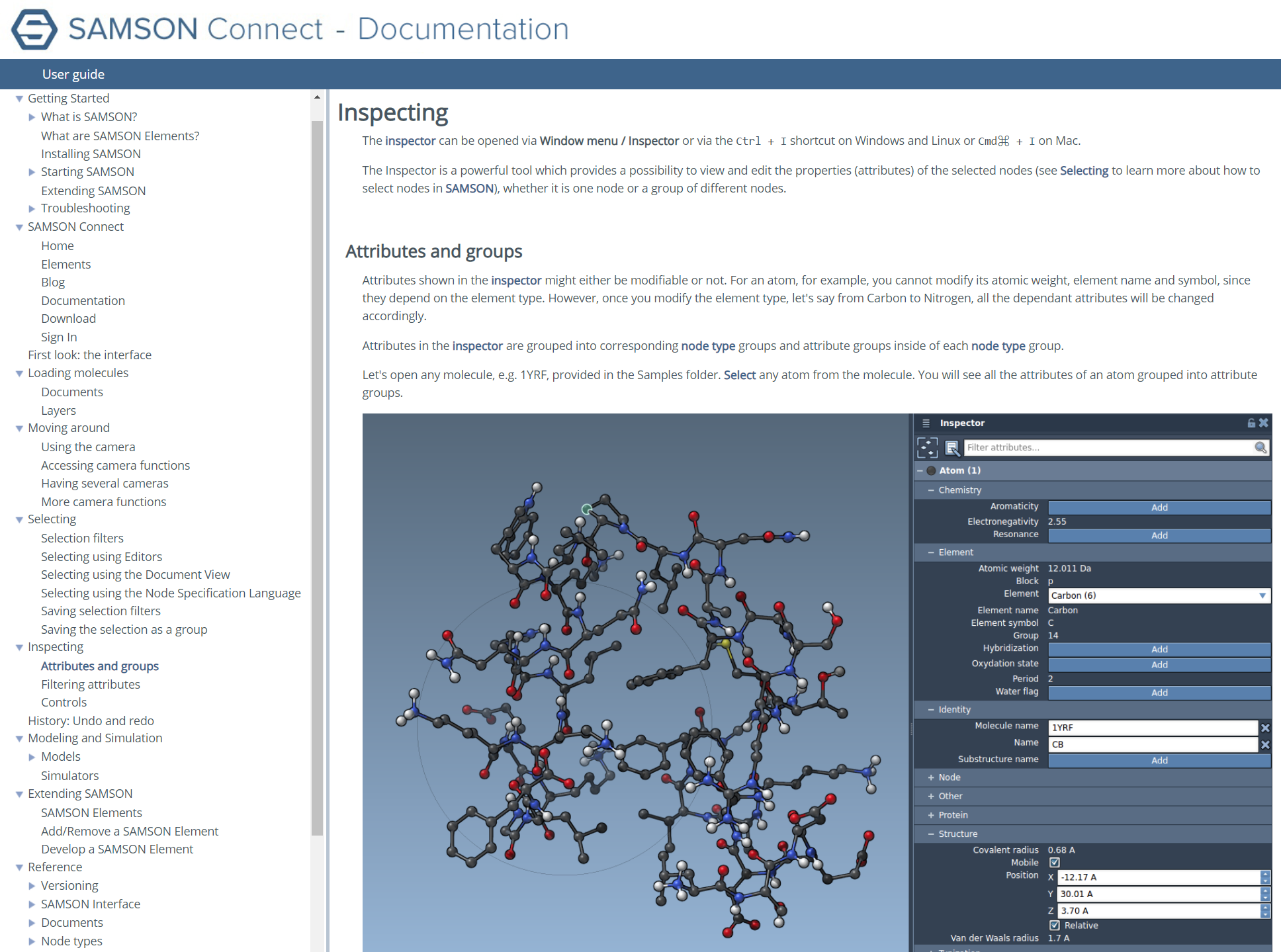
Ask the Doc
If you need more help, head to the extensive Documentation Center
for complete guides and tutorials.
Learn about the most advanced features and become a SAMSON master.
Learn more.
Learn about the most advanced features and become a SAMSON master.
Learn more.
A platform for developers,
too
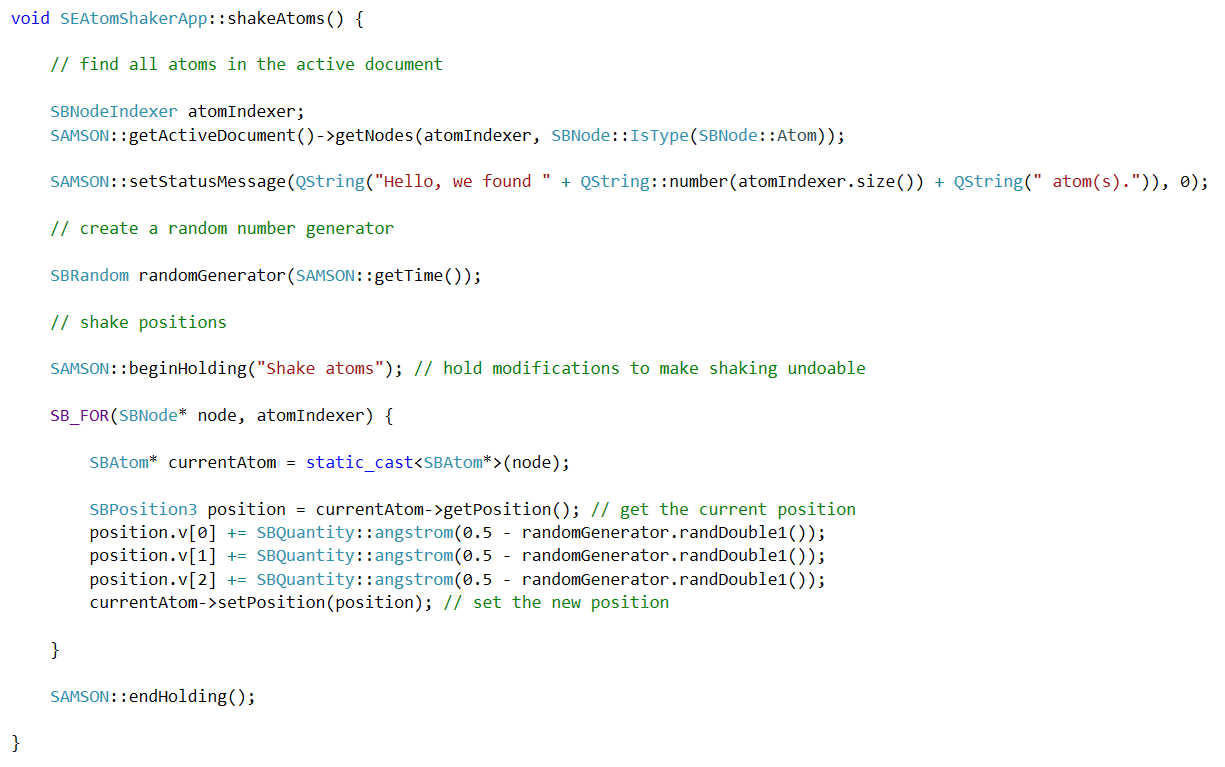
Develop
Download the SAMSON SDK and create new apps, editors, force
fields, services, etc. with maximal performance and control.
Reduce your development costs: write once and deploy everywhere with SAMSON's multiplatform SDK, and use code templates and advanced SDK features to accelerate development.
Quickly integrate what you already have or break new ground. SAMSON lets you focus on your expertise and handles visualization, data management and much more.
Make your extensions free or sell subscriptions to them, and sell access to cloud computing services from SAMSON.
Schedule a meeting with us to learn about the Partner Program.
Reduce your development costs: write once and deploy everywhere with SAMSON's multiplatform SDK, and use code templates and advanced SDK features to accelerate development.
Quickly integrate what you already have or break new ground. SAMSON lets you focus on your expertise and handles visualization, data management and much more.
Make your extensions free or sell subscriptions to them, and sell access to cloud computing services from SAMSON.
Schedule a meeting with us to learn about the Partner Program.